# A tibble: 6 × 11
religion `<$10k` `$10-20k` `$20-30k` `$30-40k` `$40-50k` `$50-75k` `$75-100k`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Agnostic 27 34 60 81 76 137 122
2 Atheist 12 27 37 52 35 70 73
3 Buddhist 27 21 30 34 33 58 62
4 Catholic 418 617 732 670 638 1116 949
5 Don’t kn… 15 14 15 11 10 35 21
6 Evangeli… 575 869 1064 982 881 1486 949
# ℹ 3 more variables: `$100-150k` <dbl>, `>150k` <dbl>,
# `Don't know/refused` <dbl>Joins data frames and pivoting
2025-12-01
Housekeeping
Content today is optional; might be useful for your final project!
Pivoting
Wide data
Let’s take a look at first few observations of the relig_income data frame from tidyverse package:
This data is currently in “wide” format: a row has more than one observation. That is, the same outcome variable appears in multiple columns.
The outcome observation that spans across several columns is income range.
The different incomes columns are essentially different levels of the same categorical variable.
Long data
We might want long format: the outcome variable only exists in one column, and a second column/variable tells us the different levels.
Each row has one observation, but the units of observation (in this case, number of people in each income bracket) are repeated down one column.
This is helpful for us to perform group_by() or facet_wrap().
Converting from wide -> long
We will do this with the pivot_longer() function. This function requires a couple of arguments:
cols: which columns to pivot into a “longer” format. That is, the columns that we should “move”names_to: a string character that provides the new column name for the categorical variable you are creatingvalues_to: a string character that provides the new variable name for the response/outcome variable that is common across all levels of the categorical variable
pivot_longer()
Resulting data frame:
# A tibble: 20 × 3
religion income_range count
<chr> <chr> <dbl>
1 Agnostic <$10k 27
2 Agnostic $10-20k 34
3 Agnostic $20-30k 60
4 Agnostic $30-40k 81
5 Agnostic $40-50k 76
6 Agnostic $50-75k 137
7 Agnostic $75-100k 122
8 Agnostic $100-150k 109
9 Agnostic >150k 84
10 Agnostic Don't know/refused 96
11 Atheist <$10k 12
12 Atheist $10-20k 27
13 Atheist $20-30k 37
14 Atheist $30-40k 52
15 Atheist $40-50k 35
16 Atheist $50-75k 70
17 Atheist $75-100k 73
18 Atheist $100-150k 59
19 Atheist >150k 74
20 Atheist Don't know/refused 76Converting long -> wide
Let’s look at the fish_encounters data. Convince yourself that it’s currently in long format!
# A tibble: 6 × 3
fish station seen
<fct> <fct> <int>
1 4842 Release 1
2 4842 I80_1 1
3 4842 Lisbon 1
4 4842 Rstr 1
5 4842 Base_TD 1
6 4842 BCE 1- We want to pivot the data such that each fish is an observation, and we can easily see which stations it was observed at.
- We can do this using the
pivot_wider()function!
pivot_wider()
The pivot_wider() function takes two arguments:
names_from: the name of the variable(s) in the data frame to get the name of the output column.values_from: the name of the variable(s) in the data frame to get the cell values from
The input to
names_fromshould be the categorical variable(s), and the different levels of this categorical variable will be the new column names.- What should we fill these cells with? The values specified by
values_from.
- What should we fill these cells with? The values specified by
pivot_wider()
# A tibble: 19 × 12
fish Release I80_1 Lisbon Rstr Base_TD BCE BCW BCE2 BCW2 MAE MAW
<fct> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
1 4842 1 1 1 1 1 1 1 1 1 1 1
2 4843 1 1 1 1 1 1 1 1 1 1 1
3 4844 1 1 1 1 1 1 1 1 1 1 1
4 4845 1 1 1 1 1 NA NA NA NA NA NA
5 4847 1 1 1 NA NA NA NA NA NA NA NA
6 4848 1 1 1 1 NA NA NA NA NA NA NA
7 4849 1 1 NA NA NA NA NA NA NA NA NA
8 4850 1 1 NA 1 1 1 1 NA NA NA NA
9 4851 1 1 NA NA NA NA NA NA NA NA NA
10 4854 1 1 NA NA NA NA NA NA NA NA NA
11 4855 1 1 1 1 1 NA NA NA NA NA NA
12 4857 1 1 1 1 1 1 1 1 1 NA NA
13 4858 1 1 1 1 1 1 1 1 1 1 1
14 4859 1 1 1 1 1 NA NA NA NA NA NA
15 4861 1 1 1 1 1 1 1 1 1 1 1
16 4862 1 1 1 1 1 1 1 1 1 NA NA
17 4863 1 1 NA NA NA NA NA NA NA NA NA
18 4864 1 1 NA NA NA NA NA NA NA NA NA
19 4865 1 1 1 NA NA NA NA NA NA NA NA- Note that there are some
NAvalues after pivoting. From the Help file, this is because misses were not directly recorded in the original form of the data. Try adding the argumentvalues_fill = 0to thepivot_wider()function.
Joins
Joining data frames
Assume we have two data frames, df1 and df2. There are some shared variable(s) across the two.
We want to combine df1 and df2 into one single data frame. The dplyr code will look something like:
How do we combine
df1anddf2them? The following mutating joins give us rules:left_join(): keep all cases fromdf1right_join(): keep all cases fromdf2full_join(): keep all cases from bothdf1anddf2inner_join(): keep all cases fromdf1where there are matching values indf2(more complicated in the scenario of multiple matches)
I know this is confusing! Examples will help clarify!
Setup
For the next few slides…
I recommend writing these two down, as we will work with them in the next few slides.
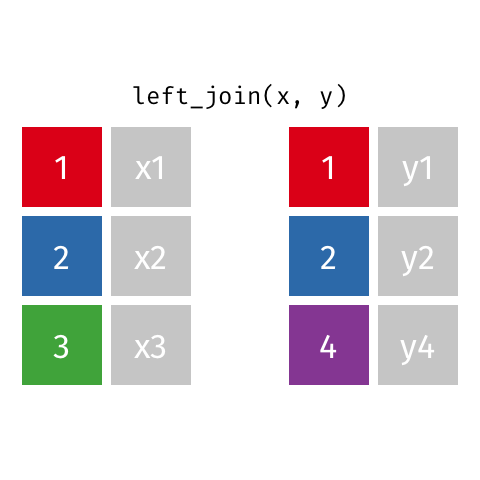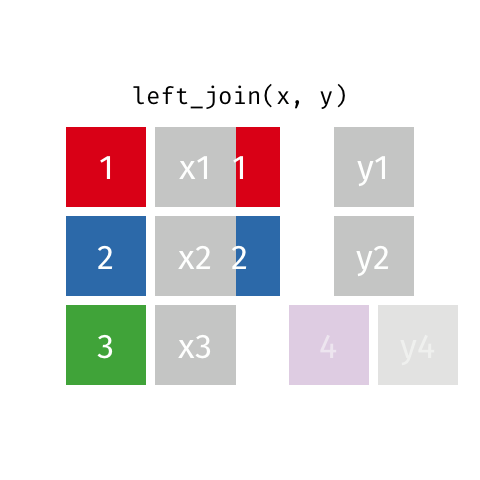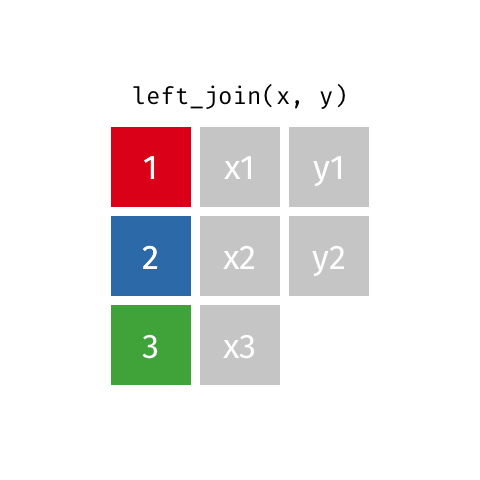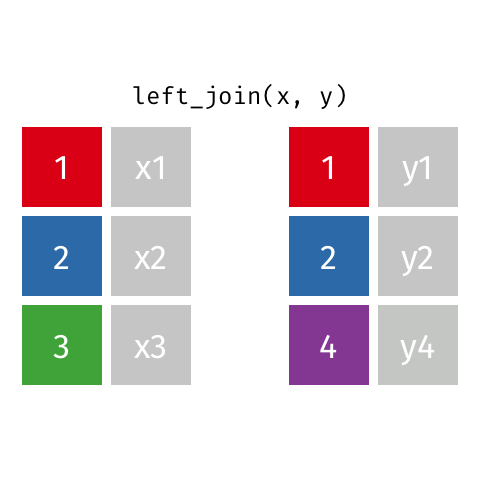
Note! The variable ID is shared between these two data frames. We will “join on” ID.
left_join(df1, df2)
Adds columns to df1 from df2, matching the cases in df1. Retains ALL cases from left data frame df1, and adds NA for variables from df2 that have no match.
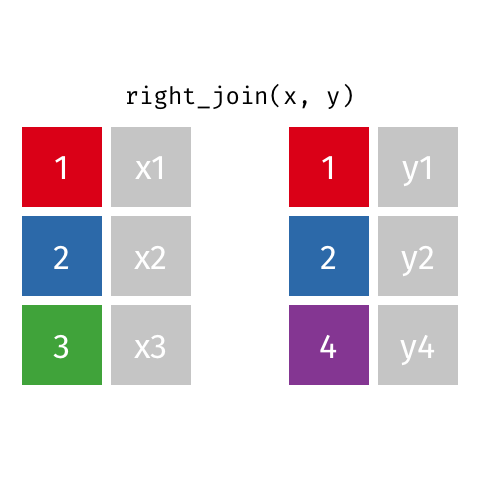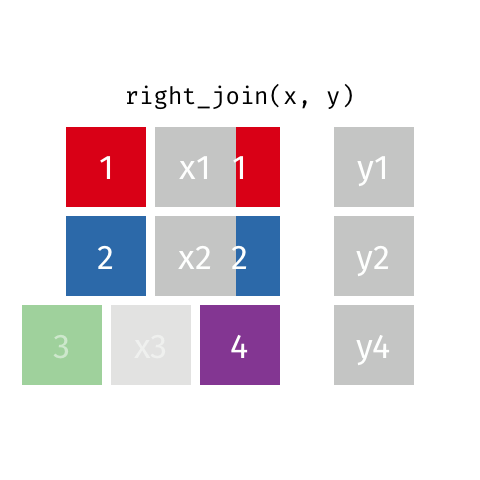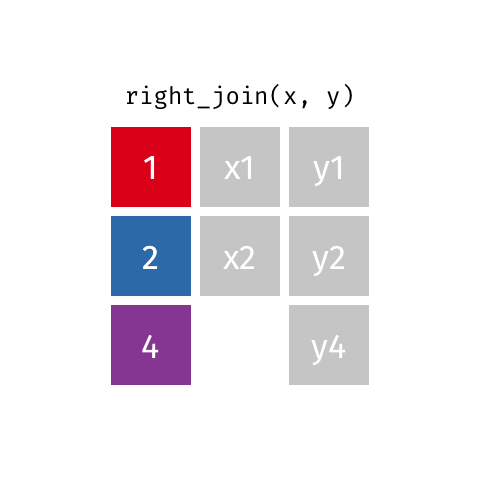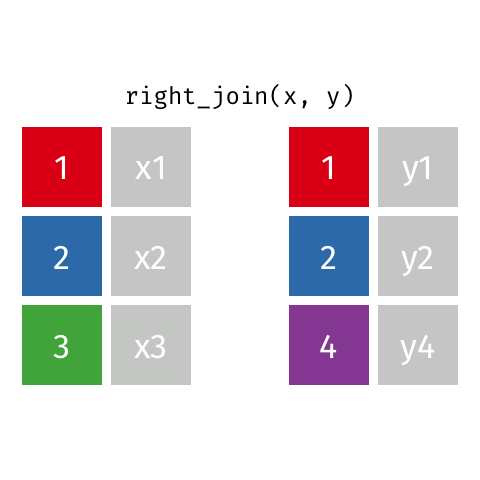
right_join(df1, df2)
Adds columns to df1 from df2, matching the cases in df2. Retains ALL cases from right data frame df2, and adds NA for variables from df1 that have no match.
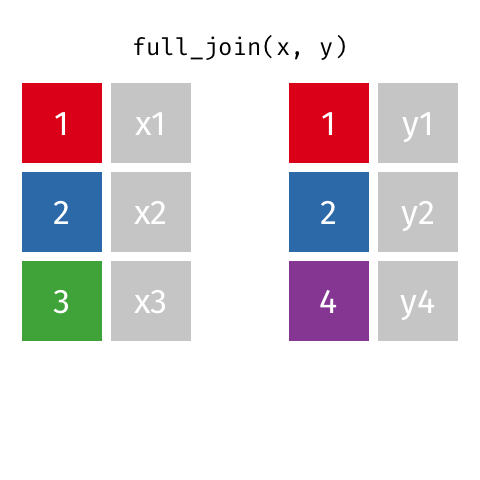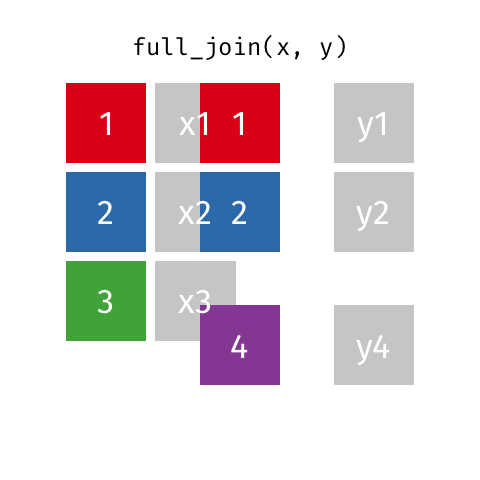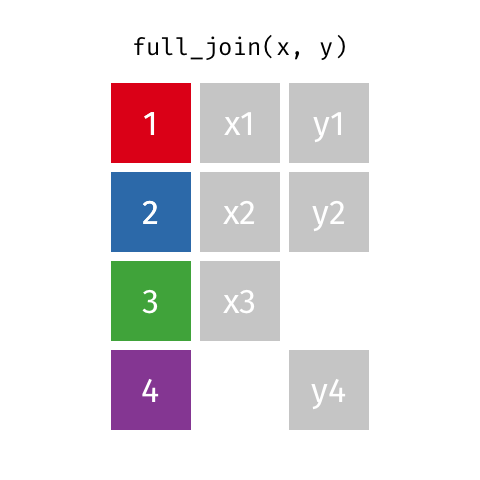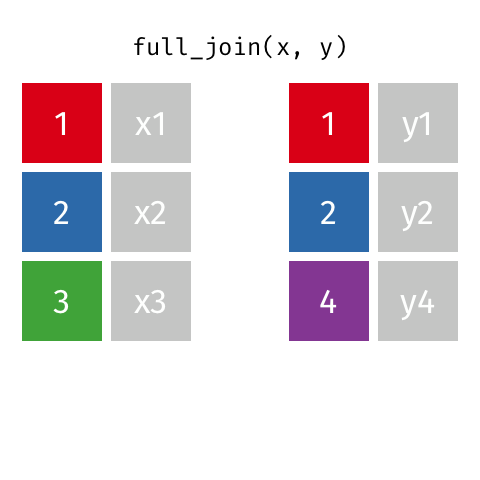
full_join(df1, df2)
Adds columns to df1 from df2, fully combining the two data frames, matching when possible. Retains ALL cases from df1 AND df2, and adds NAs as necesary.
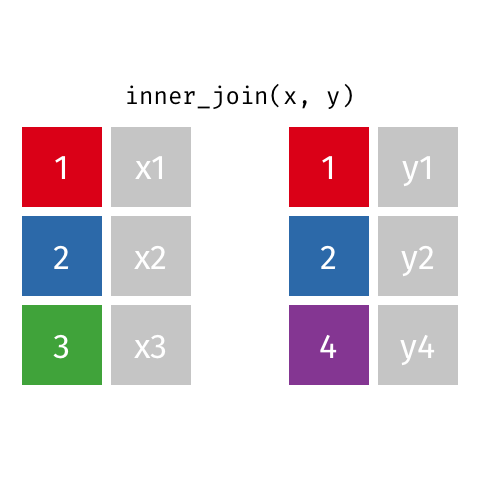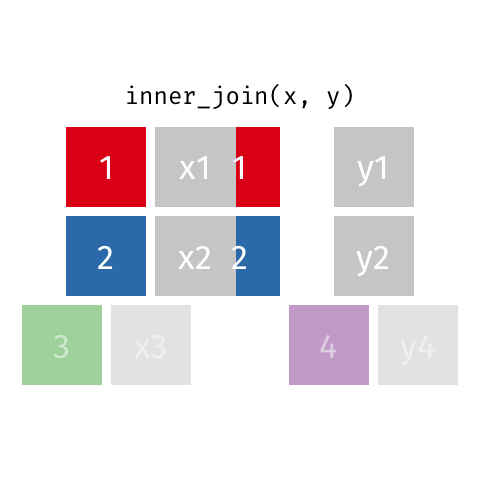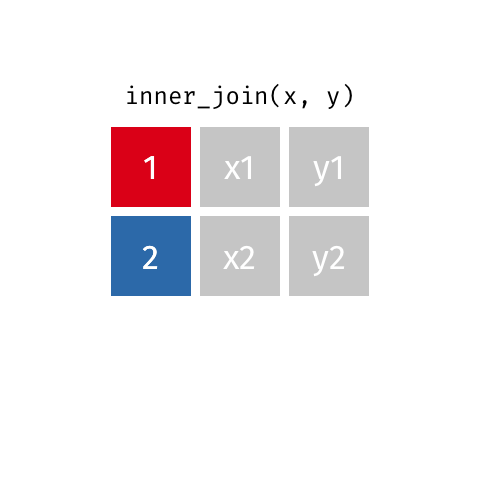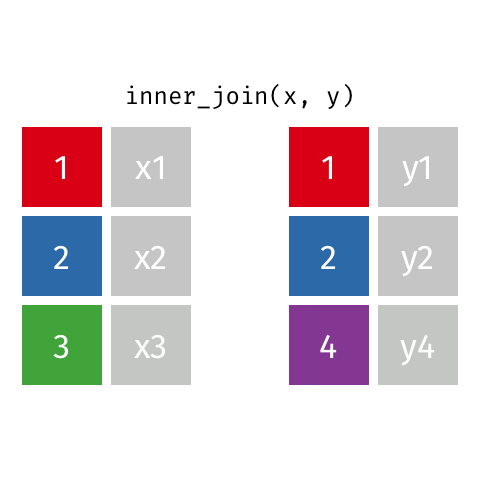
inner_join(df1, df2)
Adds columns to df1 from df2, retaining only cases that exist in both df1 AND df2.
Comprehension check
- In which of the joins are
NAs possibly introduced? In which of the joins areNAs never going to be introduced? - If we have some
df1anddf2, in what scenario will the result of the four joins be the same? (i.e. what has to be true aboutdf1anddf2?) - What do you think we call these “mutating” joins? Think about where we’ve seen that verb before!
- Note: it’s possible to join on more than one variable!
Example
We have data on fishery harvests (in tons) by countries from 2016, stored in data frame fish:
| country | capture | aquaculture |
|---|---|---|
| Afghanistan | 1000 | 1200 |
| Albania | 7886 | 950 |
| Algeria | 95000 | 1361 |
| American Samoa | 3047 | 20 |
| Andorra | 0 | 0 |
| Angola | 486490 | 655 |
| Antigua and Barbuda | 3000 | 10 |
| Argentina | 755226 | 3673 |
Suppose I would like to explore the data on a continent level. We have another data frame called continent that looks like this:
| Country | continent |
|---|---|
| Afghanistan | Asia |
| Åland Islands | Europe |
| Albania | Europe |
| Algeria | Africa |
| American Samoa | Oceania |
| Andorra | Europe |
| Angola | Africa |
| Anguilla | Americas |
Discuss
fish data frame snapshot:
| country | capture | aquaculture |
|---|---|---|
| Afghanistan | 1000 | 1200 |
| Albania | 7886 | 950 |
| Algeria | 95000 | 1361 |
| American Samoa | 3047 | 20 |
| Andorra | 0 | 0 |
continents data frame snapshot:
| Country | continent |
|---|---|
| Afghanistan | Asia |
| Åland Islands | Europe |
| Albania | Europe |
| Algeria | Africa |
| American Samoa | Oceania |
We want to keep all rows and columns from
fishand add a column for corresponding continents. Which join function should we use?We want to keep only the rows from
fishfor which we have a corresponding continent and add a column for corresponding continents. Which join function should we use?Which variable(s) are we joining on?
Example (cont.)
Question 1:
| country | capture | aquaculture | continent |
|---|---|---|---|
| Afghanistan | 1000 | 1200 | Asia |
| Albania | 7886 | 950 | Europe |
| Algeria | 95000 | 1361 | Africa |
| American Samoa | 3047 | 20 | Oceania |
| Andorra | 0 | 0 | Europe |
| Angola | 486490 | 655 | Africa |
| Antigua and Barbuda | 3000 | 10 | NA |
| Argentina | 755226 | 3673 | Americas |
- Notice the
NA
Question 2:
| country | capture | aquaculture | continent |
|---|---|---|---|
| Afghanistan | 1000 | 1200 | Asia |
| Albania | 7886 | 950 | Europe |
| Algeria | 95000 | 1361 | Africa |
| American Samoa | 3047 | 20 | Oceania |
| Andorra | 0 | 0 | Europe |
| Angola | 486490 | 655 | Africa |
| Argentina | 755226 | 3673 | Americas |
| Armenia | 3758 | 16381 | Asia |
- Notice we don’t have two copies of same
country/Countryvariable!
Piping into xxx_join()
In the following line of code, when would an NA be introduced? (Answer forthcoming.)
Remember in data wrangling, we pipe from a previous line of code directly into another
We can pipe directly into the join functions! The following achieves the same as above:
Answer to previous question
NAs would be introduced if there is a country in continents that does not appear in fish. See the last 50-ish cases: